
Estimating County-Level Mortality Rates Using Highly Censored Data From CDC WONDER
ORIGINAL RESEARCH — Volume 16 — June 13, 2019
This article is part of the Population Health, Place, and Space: Spatial Perspectives in Chronic Disease Research and Practice collection.
Harrison Quick, PhD
Suggested citation for this article: Quick H. Estimating County-Level Mortality Rates Using Highly Censored Data From CDC WONDER. Prev Chronic Dis 2019;16:180441. DOI: http://dx.doi.org/10.5888/pcd16.180441.
PEER REVIEWED
Summary
What is already known on this topic?
Ignoring the impact of suppression due to small counts leads to biased inference.
What is added by this report?
This work describes and compares multiple approaches for analyzing highly suppressed data from CDC WONDER. R and WinBUGS code are provided to conduct the analyses.
What are the implications for public health practice?
The use of spatial Bayesian models can yield improved inference from the analysis of highly suppressed data such as those available on CDC WONDER.
Abstract
Introduction
CDC WONDER is a system developed to promote information-driven decision making and provide access to detailed public health information to the general public. Although CDC WONDER contains a wealth of data, any counts fewer than 10 are suppressed for confidentiality reasons, resulting in left-censored data. The objective of this analysis was to describe methods for the analysis of highly censored data.
Methods
A substitution approach was compared with 1) a simple, nonspatial Bayesian model that smooths rates toward their statewide averages and 2) a more complex Bayesian model that accounts for spatial and between-age sources of dependence. Age group–specific county-level data on heart disease mortality were used for the comparisons.
Results
Although the substitution and nonspatial approach provided age-standardized rate estimates that were more highly correlated with the true rate estimates, the estimates from the spatial Bayesian model provided a superior compromise between goodness-of-fit and model complexity, as measured by the deviance information criterion. In addition, the spatial Bayesian model provided rate estimates with greater precision than the nonspatial approach; in contrast, the substitution approach did not provide estimates of uncertainty.
Conclusion
Because of the ability to account for multiple sources of dependence and the flexibility to include covariate information, the use of spatial Bayesian models should be considered when analyzing highly censored data from CDC WONDER.
Introduction
CDC WONDER (Wide-ranging ONline Data for Epidemiologic Research) is a system developed by the Centers for Disease Control and Prevention (CDC) to promote information-driven decision making by public health practitioners and researchers and provide access to detailed public health information to the general public (1). Although CDC WONDER contains a wealth of data, it has limitations. Per CDC policy (2), any counts fewer than 10 should be suppressed for confidentiality reasons, resulting in left-censored data. Because of high rates of suppression, many chronic disease researchers opt to focus their inference in a few highly populated regions (3) or state- or national-level trends (4), despite known geographic disparities in many chronic disease outcomes (5,6). This suppression may also discourage research on disparities between subsets of the population (eg, race or sex disparities) to avoid reducing already small counts below suppression thresholds. In short, suppression of small counts exacerbates many issues commonly encountered in the field of small area estimation, where the term “small area” refers to a geographic scale (eg, county, census tract) at which the observed data alone do not provide reliable inference. Thus, when CDC WONDER data are used to conduct surveillance, the ability to estimate rates for rural areas and minority populations — where the chronic disease burden is high (7) — is significantly hindered by data suppression.
To address CDC WONDER’s data suppression issue, Tiwari et al (8) proposed an algorithm for estimating age-standardized rates in which suppressed age-specific counts are replaced with estimates based on the county’s age-specific population size and the state-wide average rate for that age-group. For example, suppose yik denotes the number of deaths from age-bracket k in county i of a population of size nik and our inferential interest lies in λik, the corresponding mortality rate. Tiwari et al (8) proposed replacing the suppressed yik <10 with , where si denotes the state that county i belongs to and denotes the state-wide average rate for age-bracket k in state si such that
Although this approach may yield reasonable estimates, it has drawbacks. First and foremost, estimating the uncertainty in age-standardized rate estimates is not an exact science when the data are known (9,10), much less when the data are highly suppressed. Furthermore, the algorithm is not designed to account for heterogeneity in demographic information such as the racial/ethnic make-up and socioeconomic status of the counties’ populations. As a result, inference based on these substituted data may be both biased (ie, smoothing toward the wrong values) and too precise (ignoring the uncertainty due to data suppression).
When the goal is to assess geographic disparities in age-standardized rates between regions, overcoming the privacy protections to obtain trustworthy estimates of the age-specific rates and their levels of uncertainty is only half the battle. For instance, Fay (11) followed the work of Fay and Feuer (9) to construct interval estimates for ratios based on F distributions. Tiwari et al (10) modified this work to yield more efficient interval estimation for rates and ratios of rates from nonnested regions, work that was later extended by Tiwari et al (12) for when one subregion is nested within a larger region (eg, a county nested within a state); Zhu et al (13) extended these approaches to more accurately account for spatial autocorrelation. When the age-standardized rates must be estimated from suppressed data, further modifications must be made or these approaches will fail to adequately account for all sources of uncertainty, yielding interval estimates that may be too narrow (14,15).
Rather than develop the statistical theory to accurately account for substitution-based approaches to overcome CDC WONDER’s privacy restrictions in variance calculations, we consider the use of Bayesian statistical models, which rely on data augmentation to make inference on the suppressed counts. As described by Fridley and Dixon (14), data augmentation approaches estimate the suppressed counts via multiple imputation (16) while simultaneously making inference on the parameters of interest — for example, λik and the effects of potential risk factors. As noted by Zhu et al (13), Bayesian methods for modeling spatial data (17) can yield improved rate estimates when data are limited while simultaneously providing a mechanism for estimating uncertainty in rate estimates — uncertainty that can be seamlessly propagated into estimates such as age-standardized rates and rate ratios. That said, a key drawback of Bayesian methods is their tendency to rely on computationally burdensome Markov chain Monte Carlo (MCMC) methods.
The objective of this analysis was to illustrate 2 Bayesian approaches for estimating county-level mortality rates, by using heart disease mortality data from 1980 obtained from CDC WONDER (18), and to compare these results with those generated by the approach of Tiwari et al (8). In particular, we used a simple, nonspatial Bayesian model, which produces estimates similar to those from Tiwari et al (8), along with a more complex Bayesian model that accounts for spatial and between-age sources of dependence.
Methods
The study population for this analysis included all residents of the contiguous United States aged 35 or older during 1980. These data have multiple advantages. Because these data were collected before CDC’s suppression guidelines (2) went into effect, the public-use data are complete and free of suppression. Furthermore, because county definitions changed in several ways during the 1980s, the choice of data from 1980 allowed use of readily available shapefiles from the US Census Bureau for the I = 3,109 counties (or county equivalents) in the contiguous United States. To replicate the analysis of Tiwari et al (8), the data were separated into K = 6 groups: those aged 35 to 44, 45 to 54, 55 to 64, 65 to 74, 75 to 84, and 85 or older. Annual counts of heart disease–related deaths per county per age-group were obtained via CDC WONDER (18) and were defined as those for which the underlying cause of death was “diseases of the heart” according to the International Classification of Diseases, Ninth Revision (codes 390–398, 402, 404–429). Of the more than 18,000 counts in this data set, nearly half were fewer than 10.
Statistical model
Recall that yik and nik denote the number of deaths and the population size in age group k in county i. To model these data, we considered 2 approaches: a simple Poisson-gamma model and a multivariate spatial Bayesian model. Although the former illustrates how a Bayesian model with weakly informative priors can produce estimates similar to those obtained directly from the raw data — but with accurate uncertainty measures — the latter illustrates how Bayesian models can incorporate complex dependence structures to produce more reliable estimates. A formal definition of what constitutes a “reliable” rate and the implications of this definition are provided in the Web Appendix (https://sites.google.com/site/harryq/wonder). Because of the complexity of Bayesian models, the Web Appendix also provides technical details on the methods described in this article and includes R (19) and WinBUGS (20) code.
Poisson-gamma model
Following the advice of Brillinger (21), we assumed
for i = 1, . . ., I and k = 1, …, K. Because we wished to fit Equation 2 using a Bayesian framework, we had to specify a prior distribution for each λik. A convenient choice was to let
As described in the Web Appendix, can be interpreted as the prior number of events and as the prior population size, thereby providing a mechanism for comparing the informativeness of the prior to the amount of information contained in the data. For example, a prior with = 1,000 would contain the same amount of information as the data when nik = 1,000, and the posterior mean would be equal to the average of (the estimate from the prior) and (the estimate from the data). Here, we can take an empirical Bayesian approach by letting from Equation 1 and defining the informativeness of the prior to be such that 6 for all states under the restriction that the parameters respect the age distribution in the United States. To better accommodate low rates among the younger age groups, which produce a preponderance of zero counts, we modified the prior in Equation 3 based on the suggestion of Kerman (22) by letting
This prior specification can be considered relatively noninformative because 96.4% of US counties had more than 8 heart disease–related deaths in 1980. A more complete discussion of this model is provided in the Web Appendix.
Multivariate conditional autoregressive model
Although the prior specification in Equation 4 is a convenient choice, it does not take full advantage of the possibilities of Bayesian modeling. In particular, Equation 4 does not account for spatial relationships or the relationships between different age groups. To allow for such structures to be included in the model, we considered Poisson regression models, where
Here, xik denotes a vector of county-specific covariates with corresponding age-specific regression coefficients, βk; for example, including state-level effects could help account for important health policy differences across state lines. For this analysis, we simply assumed ; that is, a model with age-specific intercept parameters. To account for spatial and between-age sources of dependence, we first followed the approach of Besag et al (17) and defined , where zik accounts for spatial structure within each age-group and denotes an exchangeable (ie, nonspatial) random effect. More specifically, the conditional autoregressive (CAR) model of Besag et al (17) imposes spatial structure by shrinking each zik toward the values in neighboring counties (ie, counties that share a border), where the strength of this shrinkage is controlled by the number of neighboring counties.
Although the CAR model is a powerful tool for analyzing spatial data, it does not account for possible correlation between the multiple age groups. To account for this, we instead considered a multivariate extension of the CAR model: the multivariate CAR (MCAR) model of Gelfand and Vounatsou (23). As with the CAR model, the MCAR shrinks estimates toward their neighboring values; unlike the CAR model, however, the MCAR explicitly models the between-group correlation in the data and leverages these correlations to produce more precise age-specific rate estimates. MCAR models were used recently to model spatially referenced survival times in cancer data (24), temporal trends in county-level asthma hospitalization rates (25), temporal trends in heart disease mortality by race and sex (26), and temporal trends in age-specific stroke mortality (27), among many other applications. Full details, including a discussion of the prior distributions used, are provided in the Web Appendix.
Bayesian inference
Fitting the models in Equation 4 or Equation 5 while accounting for the suppression of counts fewer than 10 requires the use of MCMC algorithms. Because of the reliance on MCMC, inference from these Bayesian models is based on samples generated from the posterior distribution — for example, for l = 1, …, L, where L denotes the number of samples. These samples can then be used to compute quantities such as the age-standardized mortality rate:
where πk denotes a prespecified standard age distribution (eg, based on the 2010 US standard population). To summarize the posterior distribution, it is common to use the posterior median and the 95% credible interval (constructed from the 2.5 and 97.5 percentiles of the posterior samples and analogous to classical 95% confidence intervals).
Comparison of approaches
To compare the various estimation approaches, we first considered simple correlations between the estimates and the rates obtained from the complete data (as considered by Tiwari et al [8]) and correlations between the age-standardized rates and the age-specific rates. The goal of these comparisons was not to demonstrate whether one approach is superior to another but rather to demonstrate the degree to which the approaches are similar to one another. In addition, we also compared the 2 Bayesian approaches by using the deviance information criterion (DIC) (28), which uses the posterior samples to produce a measure that is a compromise between model fit (denoted by ) the effective number of parameters in the model. Additional details on DIC, including a discussion of its use with censored data, are provided in the Web Appendix.
Creation of maps
Maps were created by using the R statistical software (The R Foundation). Code is available in step 6 of the walkthrough in the Web Appendix (https://sites.google.com/site/harryq/wonder).
Results
The maps of the age-standardized rates generated from the raw data (Figure 1A) and the maps generated by the Poisson-gamma model (Figure 1C) have strong similarities, while artifacts of substituting state-wide averages for suppressed counts based on the approach of Tiwari et al (8) lead to elevated estimates in many rural counties in the upper Midwest (Figure 1B). In contrast, the map of the estimates from the MCAR model (Figure 1D) preserves the overall trends in the data while producing significantly smoother rate estimates.
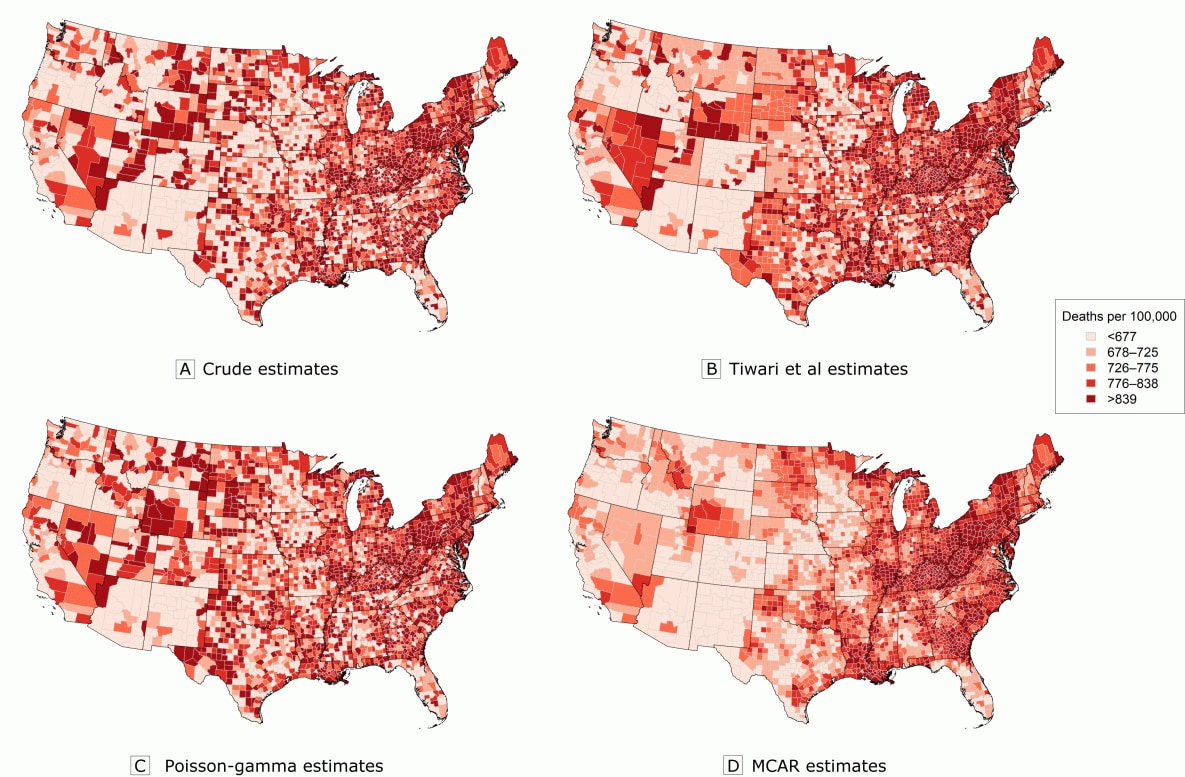
Figure 1.
Estimates of age-standardized heart disease mortality rates from 1980. A, Crude age-standardized rates based solely on the data. B, Estimates obtained by using the approach of Tiwari et al (8). C, Estimated posterior medians from the Poisson-gamma model. D, Estimated posterior medians from the multivariate conditional autoregressive model (MCAR). Data source: Centers for Disease Control and Prevention (18). [A text version of this figure is also available.]
Estimates of age-standardized heart disease mortality rates from 1980. A, Crude age-standardized rates based solely on the data. B, Estimates obtained by using the approach of Tiwari et al (8). C, Estimated posterior medians from the Poisson-gamma model. D, Estimated posterior medians from the multivariate conditional autoregressive model (MCAR). Data source: Centers for Disease Control and Prevention (18). [A text version of this figure is also available.]
The correlation results (Table 1) largely support this assessment. The Poisson-gamma approach produced age-standardized rate estimates that were the most highly correlated with the true rates, although the estimates obtained by using the substitution approach of Tiwari et al (8) had nearly an identical correlation. These 2 approaches differed in age-specific rate estimates. In particular, although the Poisson-gamma approach appeared to struggle for adults aged 35 to 44 — producing estimates that were less correlated with the truth — it outperformed the substitution approach for all groups aged 55 or older. Figure 2, which displays the age-specific rate estimates for adults aged 35 to 44 and adults 85 or older, explains how this occurred. Here, although the approach of Tiwari et al (8) gave every suppressed county in each state the same rate (by design), the Poisson-gamma model tended to overestimate rate estimates for those aged 35 to 44. According to Kerman (22), this overestimation of rates when counts are very small was to be expected. Furthermore, unlike the approach of Tiwari et al (8), the Poisson-gamma model produced full posterior distributions for each age-specific rate estimate, thereby allowing quantification of the uncertainty in these estimates. (Figure B.3 in the Web Appendix illustrates how only 4.5% of estimates for those aged 35 to 44 and 42.8% of all age-specific rate estimates from the Poisson-gamma model were deemed reliable.) When estimating rates for those 85 or older, the Poisson-gamma model permitted heterogeneity within states (Figure 2E); the inability to permit such heterogeneity is a key weakness of the approach of Tiwari et al (8). Further evaluation of the low age-specific correlations is provided in the Web Appendix (Figures B.1 and B.2).
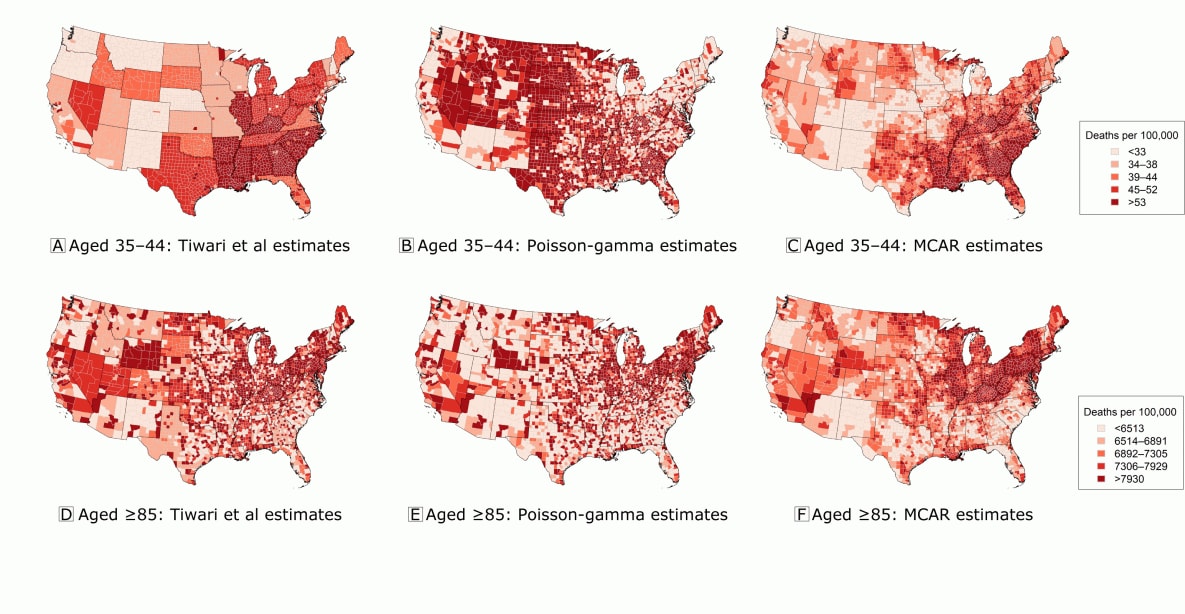
Figure 2.
Comparison of 3 approaches for estimating age-standardized heart disease mortality rates for 2 age groups (adults aged 35 to 44 and adults aged ≥85) from 1980. A, Estimates for adults aged 35 to 44 obtained by using the approach of Tiwari et al (8). B, Estimated posterior medians for adults aged 35 to 44 from the Poisson-gamma model. C, Estimated posterior medians for adults aged 35 to 44 from the multivariate conditional autoregressive model (MCAR). D, Estimates for adults aged ≥85 obtained by using the approach of Tiwari et al (8). E, Estimated posterior medians for adults aged ≥85 from the Poisson-gamma model. F, Estimated posterior medians for adults aged ≥85 from the multivariate conditional autoregressive model (MCAR). Data source: Centers for Disease Control and Prevention (18). [A text version of this figure is also available.]
Comparison of 3 approaches for estimating age-standardized heart disease mortality rates for 2 age groups (adults aged 35 to 44 and adults aged ≥85) from 1980. A, Estimates for adults aged 35 to 44 obtained by using the approach of Tiwari et al (8). B, Estimated posterior medians for adults aged 35 to 44 from the Poisson-gamma model. C, Estimated posterior medians for adults aged 35 to 44 from the multivariate conditional autoregressive model (MCAR). D, Estimates for adults aged ≥85 obtained by using the approach of Tiwari et al (8). E, Estimated posterior medians for adults aged ≥85 from the Poisson-gamma model. F, Estimated posterior medians for adults aged ≥85 from the multivariate conditional autoregressive model (MCAR). Data source: Centers for Disease Control and Prevention (18). [A text version of this figure is also available.]
Looking at the correlation results (Table 1) and the maps in Figure 1, one may wonder why we bother fitting the complex MCAR model. The DIC results (Table 2) explain why. Here, the MCAR model offered a model fit that is similar to the fit of the Poisson-gamma model (as measured by ) while doing so with far fewer “effective model parameters” (pD). To understand how this can be, recall that each λik in Equation 4 had its own independent prior distribution; that is, the Poisson-gamma model did not shrink the λik toward each other, producing estimates of the (pD) for older age groups that approach the full I = 3,109 number of parameters. In contrast, the MCAR model explicitly imposed dependence between its model parameters, resulting in estimates of the (pD) that were nearly 80% less than those from the Poisson-gamma model (eg, 10,785 vs 2,307). In addition, the estimates produced by the MCAR model were more precise (Web Appendix), and the smooth geographic patterns in Figure 1D, Figure 2C, and Figure 2F may provide clearer insight into the underlying trends in heart disease mortality.
Discussion
This analysis highlighted some of the benefits of using Bayesian methods to account for left-censored data like those encountered in CDC WONDER. Although the Poisson-gamma model is a relatively simple approach, models (such as the MCAR model) that explicitly account for multivariate spatial dependence structures can lead to better inference by leveraging other sources of information to produce more reliable estimates.
The strengths of the MCAR model described in this analysis extend beyond modeling censored data to the broader field of small area estimation. As alluded to in the discussion of Equation 5, many benefits are associated with using the MCAR model in conjunction with covariate information when modeling chronic disease outcomes. Combining covariate information with spatial structure can produce more reliable estimates of the rates themselves, which is beneficial for disease surveillance, while simultaneously conducting inference on the potential risk factors that are included as covariates. When the covariates in the analysis are themselves spatially structured, it can be unclear if the covariate is effecting change in the outcome or vice versa, or if an unmeasured spatial confounder is influencing both the covariate and the outcome. In these settings, including a spatial random effect can lead to a phenomenon referred to as “spatial confounding” (29) and increase the standard errors associated with these covariates. Although the notion of spatial confounding has historically been considered a drawback of spatial models (29), others have argued (30) that inference from such models can help protect against type 1 error (ie, incorrectly rejecting the null hypothesis).
Finally, although we analyzed age-specific heart disease mortality as an illustration, the MCAR model is also well suited for analyzing rarer event data via its ability to jointly model multiple outcomes. This analysis leveraged information from older age groups with higher death counts to produce more reliable estimates for those aged 35 to 44. Similarly, one could jointly model a chronic disease outcome for multiple race/ethnicities, exploiting the shared factors that may lead to increased rates for non-Hispanic white persons and racial/ethnic minorities alike. Alternatively, one could use MCAR models to simultaneously analyze multiple chronic disease outcomes with similar etiologies to improve the reliability of all estimates.
Although the suppression of data creates an obstacle to conducting chronic disease surveillance, Bayesian statistical methods such as those described in this analysis can overcome these challenges while also producing more reliable estimates with valid uncertainty measures. By illustrating the benefits of and providing code for their implementation, we hope to ease the burden of using Bayesian models and broaden their application to censored data sets available from sources like CDC WONDER, thereby improving the inference made from public-use data.
Author Information
Corresponding Author: Harrison Quick, PhD, Department of Epidemiology and Biostatistics, Drexel University, Philadelphia, PA 19104. Email: hsq23@drexel.edu.
.
References
- Centers for Disease Control and Prevention. CDC WONDER, 2017. http://wonder.cdc.gov. Accessed March 17, 2017.
- Centers for Disease Control and Prevention. CDC/ATSDR policy on releasing and sharing data. Manual; guide CDC-02. 2003. http://www.cdc.gov/maso/Policy/ReleasingData.pdf. Accessed June 30, 2015.
- Rust G, Zhang S, Malhotra K, Reese L, McRoy L, Baltrus P, et al. Paths to health equity: local area variation in progress toward eliminating breast cancer mortality disparities, 1990–2009. Cancer 2015;121(16):2765–74. CrossRef PubMed
- Wilmot KA, O’Flaherty M, Capewell S, Ford ES, Vaccarino V. Coronary heart disease mortality declines in the United States from 1979 through 2011: evidence for stagnation in young adults, especially women. Circulation 2015;132(11):997–1002. CrossRef PubMed
- Casper M, Kramer MR, Quick H, Schieb LJ, Vaughan AS, Greer S. Changes in the geographic patterns of heart disease mortality in the United States 1973 to 2010. Circulation 2016;133(12):1171–80. CrossRef PubMed
- Vaughan AS, Quick H, Pathak EB, Kramer MR, Casper M. Disparities in temporal and geographic patterns of declining heart disease mortality by race and sex in the United States, 1973–2010. J Am Heart Assoc 2015;4(12):e002567. CrossRef PubMed
- National Center for Health Statistics. Health, United States, 2017: with special feature on mortality. https://www.cdc.gov/nchs/data/hus/hus17.pdf. Accessed February 27, 2019.
- Tiwari C, Beyer K, Rushton G. The impact of data suppression on local mortality rates: the case of CDC WONDER. Am J Public Health 2014;104(8):1386–8. CrossRef PubMed
- Fay MP, Feuer EJ. Confidence intervals for directly standardized rates: a method based on the gamma distribution. Stat Med 1997;16(7):791–801. CrossRef PubMed
- Tiwari RC, Clegg LX, Zou Z. Efficient interval estimation for age-adjusted cancer rates. Stat Methods Med Res 2006;15(6):547–69. CrossRef PubMed
- Fay MP. Approximate confidence intervals for rate ratios from directly standardized rates with sparse data. Commun Stat Theory Methods 1999;28(9):2141–60. CrossRef
- Tiwari RC, Li Y, Zou Z. Interval estimation for ratios of correlated age-adjusted rates. J Data Sci 2010;8:471–82. PubMed
- Zhu L, Pickle LW, Pearson JB Jr. Confidence intervals for rate ratios between geographic units. Int J Health Geogr 2016;15(1):44. CrossRef PubMed
- Fridley BL, Dixon P. Data augmentation for a Bayesian spatial model involving censored observations. Environmetrics 2007;18(2):107–23. CrossRef
- Quick H, Groth C, Banerjee S, Carlin BP, Stenzel MR, Stewart PA, et al. Exploration of the use of Bayesian modeling of gradients for censored spatiotemporal data from the Exploration of the use of Bayesian modeling of gradients for censored spatiotemporal data from the Deepwater Horizon oil spill. Spat Stat 2014;9:166–79. CrossRef PubMed
- Rubin DB. Multiple imputation for nonresponse in surveys. New York (NY): John Wiley and Sons; 1987.
- Besag J, York J, Mollié A. Bayesian image restoration, with two applications in spatial statistics. Ann Inst Stat Math 1991;43(1):1–20. CrossRef
- Centers for Disease Control and Prevention, National Center for Health Statistics. Compressed mortality file 1979–1998. CDC WONDER on-line database, compiled from compressed mortality file CMF 1968–1988, series 20, no. 2A, 2000 and CMF 1989–1998, series 20, no. 2E, 2003. https://wonder.cdc.gov/controller/saved/D16/D10F745. Accessed March 3, 2017.
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna (AT): R Foundation for Statistical Computing; 2015. http://www.R-project.org/. Accessed February 27, 2019.
- Lunn DJ, Thomas A, Best N, Spiegelhalter D. WinBUGS — a Bayesian modelling framework: concepts, structure, and extensibility. Stat Comput 2000;10(4):325–37. CrossRef
- Brillinger DR. The natural variability of vital rates and associated statistics. Biometrics 1986;42(4):693–734. CrossRef PubMed
- Kerman J. Neutral noninformative and informative conjugate beta and gamma prior distributions. Electron J Stat 2011;5(0):1450–70. CrossRef
- Gelfand AE, Vounatsou P. Proper multivariate conditional autoregressive models for spatial data analysis. Biostatistics 2003;4(1):11–25. CrossRef PubMed
- Carlin BP, Banerjee S. Hierarchical multivariate CAR models for spatio-temporally correlated survival data (with discussion). In: JM Bernardo, M Bayarri, JO Berger, AP Dawid, D Heckerman, AFM Smith, and M West, editors. Bayesian statistics 7: Proceedings of the Seventh Valencia International Meeting. 45–63. Oxford (UK): Oxford Science Publications; 2003. P. 45–63.
- Quick H, Banerjee S, Carlin BP. Modeling temporal gradients in regionally aggregated California asthma hospitalization data. Ann Appl Stat 2013;7(1):154–76. CrossRef PubMed
- Quick H, Waller LA, Casper M. A multivariate space-time model for analysing county-level heart disease death rates by race and sex. J R Stat Soc 2018;67(1):291–304. CrossRef
- Quick H, Waller LA, Casper M. Multivariate spatiotemporal modeling of age-specific stroke mortality. Ann Appl Stat 2017;11(4):2165–77. CrossRef
- Spiegelhalter DJ, Best N, Carlin BP, van der Linde A. Bayesian measures of model complexity and fit (with discussion). J R Stat Soc B 2002;64(4):583–639. CrossRef
- Reich BJ, Hodges JS, Zadnik V. Effects of residual smoothing on the posterior of the fixed effects in disease-mapping models. Biometrics 2006;62(4):1197–206. CrossRef PubMed
- Hanks EM, Schliep EM, Hooten MB, Hoeting JA. Restricted spatial regression in practice: geostatistical models, confounding, and robustness under model misspecification. Environmetrics 2015;26(4):243–54. CrossRef































No hay comentarios:
Publicar un comentario