
Coronavirus is mutating with a second strain identified by scientists
The coronavirus is mutating as scientists identified a second strain, which is more aggressive and contagious.
As healthcare officials fight to curb the outbreak of coronavirus, a team of researchers from China says a new study shows that COVID-19 is mutating, with at least two types of virus circulating across countries now.
This transmission electron microscope image shows SARS-CoV-2—also known as 2019-nCoV, the virus that causes COVID-19—isolated from a patient in the U.S. Virus particles are shown emerging from the surface of cells cultured in the lab. The spikes on the outer edge of the virus particles give coronaviruses their name, crown-like. Credit: NIAID-RML
The more aggressive strain has infected about 70 percent of the people tested, while the less aggressive strain is tied to the rest of the cases.
The researchers named the aggressive strain as "L type," and the less aggressive type as "S type." The L type was more often seen in patients from Wuhan, China, where the virus first emerged in late December 2019. On the other hand, the S type is the one currently spreading in other countries. The L type strain has since declined steadily since January.
"These findings strongly support an urgent need for further immediate, comprehensive studies that combine genomic data, epidemiological data, and chart records of the clinical symptoms of patients with coronavirus disease 2019 (COVID-19)," they wrote in the paper published in the National Science Review, the journal of the Chinese Academy of Sciences.
In the study, the researchers have identified 149 mutations in the 103 genome sequences of the novel coronavirus. Health experts believed that these mutations happened recently, while 83 of the mutations are nonsynonymous, which means that it can affect and alter the amino acid sequence of a protein, resulting in a biological change in the organism.
"Our results suggest that the development of new variations in functional sites in the receptor-binding domain (RBD) of the spike seen in SARS-CoV-2 and viruses from pangolin SARSr-CoVs are likely caused by mutations and natural selection besides recombination," the researchers wrote in the study.
Further studies needed
The team from Peking University's School of Life Sciences and the Institut Pasteur of Shanghai part of the Chinese Academy of Sciences, added that human intervention might have placed more severe selective pressure on the L type of the virus, which can become more aggressive and spread rapidly. Meanwhile, the other type might have boosted in relative frequency because of a weaker selective pressure.
The scientists warned that the data basis of the study is minimal.
There is an urgent need for immediate and comprehensive studies that will combine genomic data, chart records, and epidemiological information on the clinical symptoms of patients infected with the coronavirus.
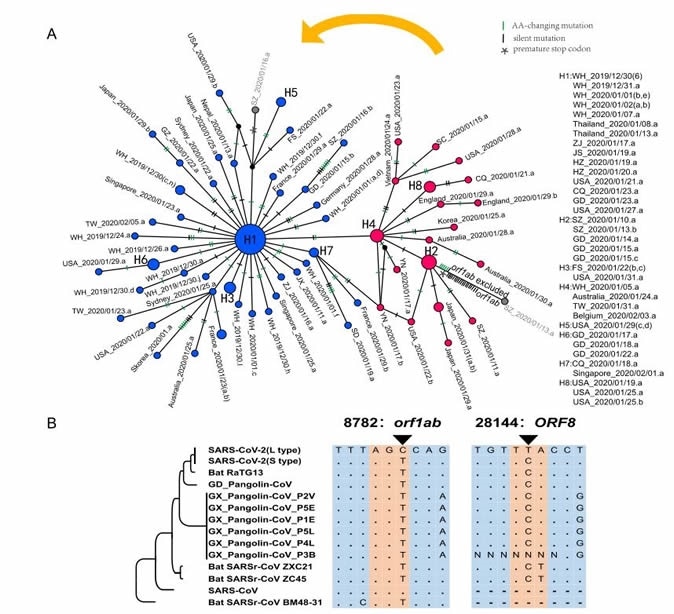
A. Haplotype analysis of SARS-CoV-2 viruses. A. The haplotype networks of SARS-CoV-2 viruses. Blue represents the L type, and red is the S type. The orange arrow indicates that the L type evolved from the S type. B. Evolution of the L and S types of SARS-CoV-2 viruses. Genome sequence alignments with the seven most closely related viruses indicated that the S type was most likely the ancient version of SARS-CoV-2.
Coronavirus by the numbers
The coronavirus cases have spiked over the past few weeks, with South Korea, Italy, and Iran gaining the spotlight over the rapid spread of the virus. The virus has spread to more than 60 countries, infecting nearly 98,000 people and killing 3,348 people, most of whom are from Wuhan City in Hubei province, China.
The World Health Organization (WHO) reports a declining supply of personal protective equipment (PPE) for healthcare workers. It urges suppliers and manufacturers to boos supply to help doctors and nurses who are working in the front line of the health crisis.
Italy and Iran have reported a sudden surge in deaths related to COVID-19. Italy has 3,858 cases and 148 deaths, while Iran has a staggering 3,513 cases and 107 deaths. South Korea has the most number of cases outside mainland China with more than 6,088 confirmed cases and 35 deaths.
Other European countries have reported their first coronavirus cases, suggesting a vast spread of the virus. The WHO has not yet declared the coronavirus outbreak a pandemic, though it said that the increase in transmission and infections is alarming.
Sources:
- World Health Organization (WHO). (2020). Coronavirus disease 2019 (COVID-19) Situation Report – 44. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200304-sitrep-44-covid-19.pdf?sfvrsn=783b4c9d_2
- Coronavirus COVID-19 Global Cases by Johns Hopkins CSSE - https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6
Journal reference:
Xiaolu Tang, Changcheng Wu, Xiang Li, Yuhe Song, Xinmin Yao, Xinkai Wu, Yuange Duan, Hong Zhang, Yirong Wang, Zhaohui Qian, Jie Cui, Jian Lu, On the origin and continuing evolution of SARS-CoV-2, National Science Review, nwaa036, https://doi.org/10.1093/nsr/nwaa036
































No hay comentarios:
Publicar un comentario