Featured article: Randomized SMILES strings improve the quality of molecular generative models
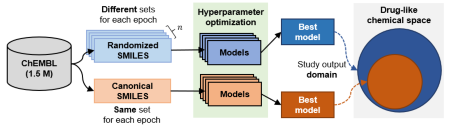
Recurrent Neural Networks (RNNs) trained with a set of molecules represented as unique (canonical) SMILES strings have shown the capacity to create large chemical spaces of valid and meaningful structures. In this article, Arús-Pous et al. performed an extensive benchmark on models trained with subsets of GDB-13 of different sizes (1 million, 10,000 and 1000), with different SMILES variants (canonical, randomized and DeepSMILES), with two different recurrent cell types (LSTM and GRU) and with different hyperparameter combinations. To guide the benchmarks, new metrics were developed that define how well a model has generalized the training set. Results show that models that use LSTM cells trained with 1 million randomized SMILES, a non-unique molecular string representation, are able to generalize to larger chemical spaces than the other approaches and they represent more accurately the target chemical space.





















.png)









No hay comentarios:
Publicar un comentario